Jerry Pelletier Laboratory
Department of Biochemistry and The Rosalind and Morris Goodman cancer Centre

Resources and Enabling tools
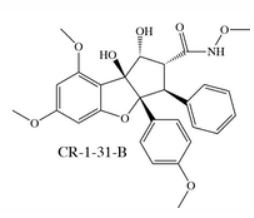
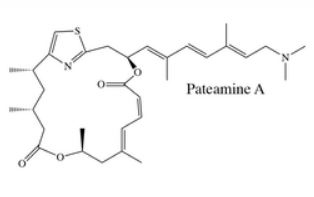
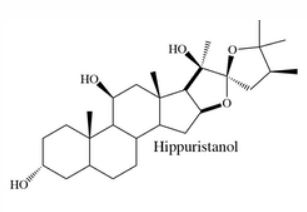
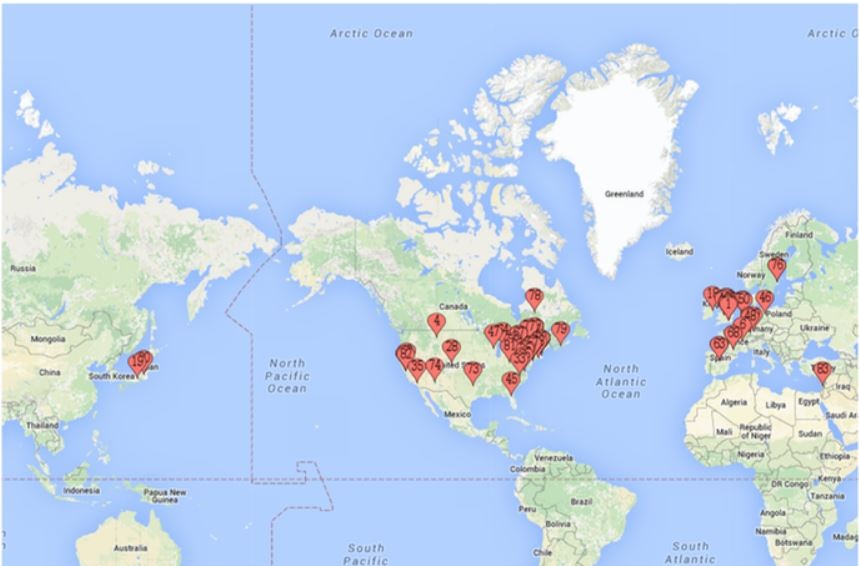
Chemical Biology Resouces. Our laboratory has amassed a unique collection of natural products, from which we identified threee potent and highly selective eIF4A inhibitors following an HTS screening campaign (PMID 16030146, 16532013, 18551192). These compounds (see below: hippuristanol, pateamine A, and rocaglates) have very different mechanisms of action (MOA). These compounds are capable of reversing resistance to several standard of care chemotherapeutics. therapies. Understanding the molecular basis for this is a current research interest of the lab. We have collaborated with many labs worldwide with these compounds - either in providing access to these unique reagents or advice on their use/effects in vitro and in vivo (see map for where these compounds have been distributed). Our laboratory has had a long standing collaboration with Dr. John Porco at Boston University (http://sites.bu.edu/porcogrp/) on work related to the rocaglates and together our efforts have afforded significant insight into the MOA of this class of natural products.
Enabling Tools for Genome Engineering. Our laboratory has been striving to improve the delivery, quality, and efficiency of CRISPR/Cas9 editing systems guided by experimental approaches. We have repurposed CRISPR/Cas9 technology for in vivo genetic screens (PMID 24298059), assessed parameters that influence Cas9 on-target versus off-target cleavage efficiency (PMID 25275497), identified small molecule modulators that favor DNA repair by homologous recombination over non-homologous end joining (PMID 26307031), and identified PAM multiplicity within target sites as being detrimental to Cas9 editing efficiency (PMID 26644285). We now use CRISPR/Cas9 on a regular basis to modify the genome to study gene function at various steps.

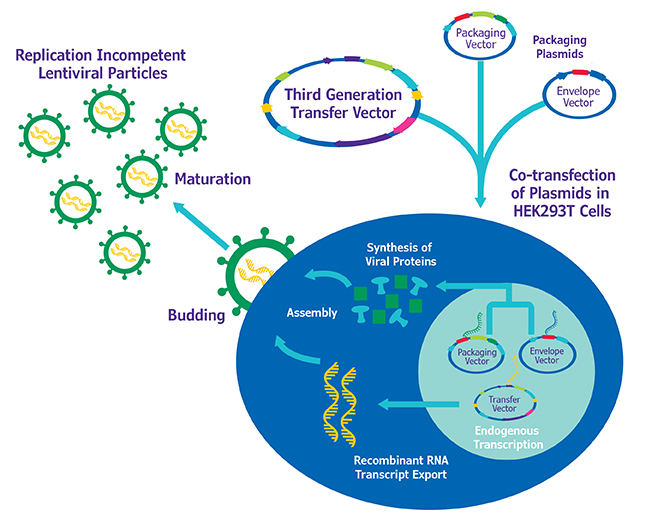
Enabling Tools for Genetic Screens. The Genetic Perturbation Core (managed by Dr. Sid Huang) houses (I) shRNA collections [human and mouse genome-wide TRC lentiviral shRNA collection (TRC1, 1.5, and 2)], (II) cDNA collections [Mission TRC3 barcoded human lentiORF genome-wide cDNA collection, MGC premier Human Lentiviral ORF Collection (Blasticidin), MGC premier Human ORFeome v8.1 and Collaboration Collections (Entry), MGC premier Mouse ORFeome Collection (Entry)] and, (III) CRISPR sgRNA collections [GeCKOv2 genome-wide, Brunello & Bri genome-wide].